HDF5 & Chunk Selection for Audio and Video
Tests on chunk selection for audio and video and its effect on access time and compression
Background on HDF5
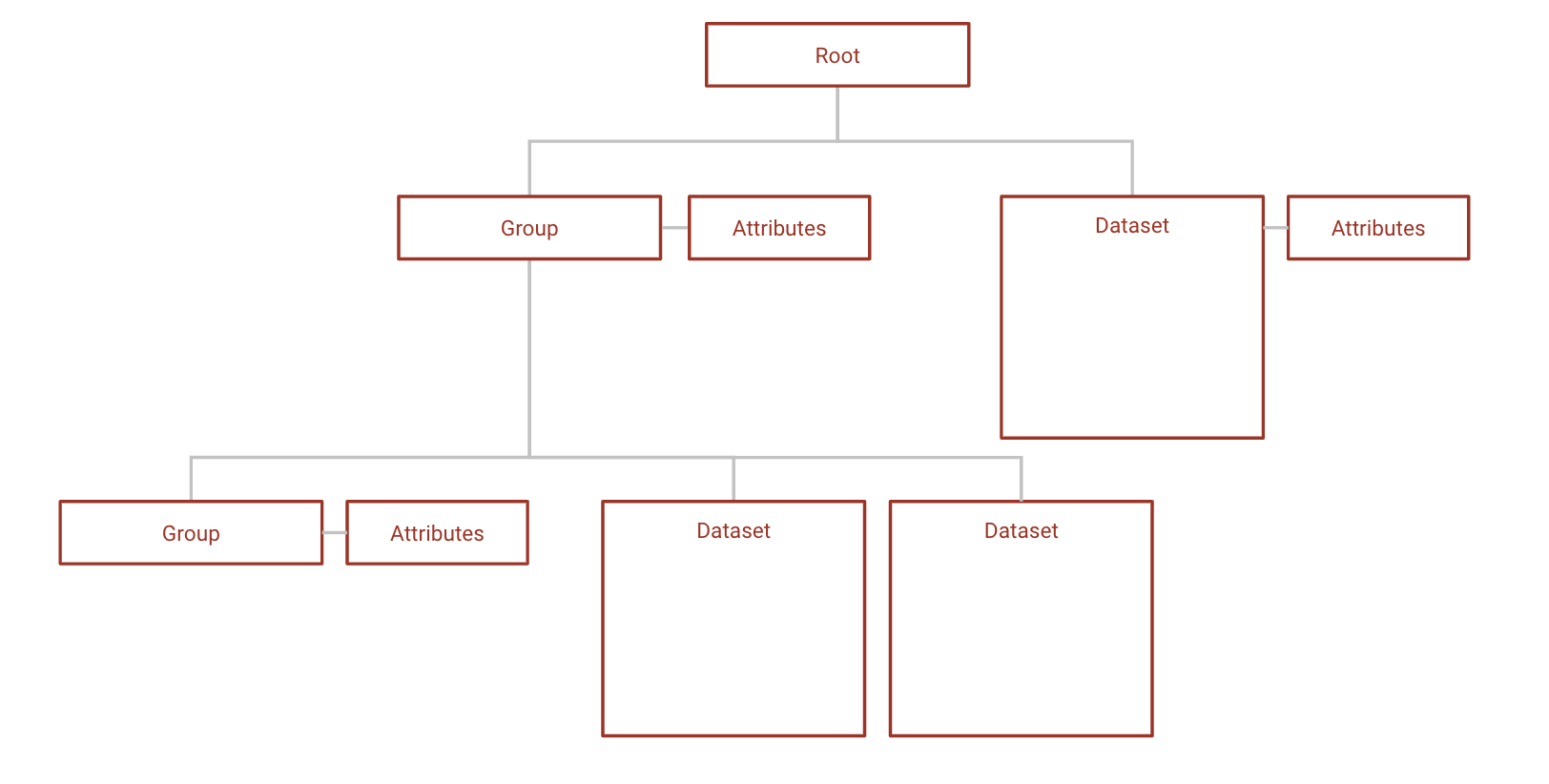
HDF5 is a file format that structures data in a similar way to a file system; data is stored in arbitrarily complex NxN datasets, which can be organized into groups and sub-groups. Groups can contain both datasets and other groups, and datasets can be of any dimension and data type. HDF5 files are also self-describing; datasets and groups both have attribute lists that store their metadata.
The HDF Group identifies why satellites use HDF5 for earth sciences data, highlighting two major benefits: customizability and scalability.
“As satellites and other instruments increase their ability to collect data at higher and higher resolution, they need a format that can scale easily and that can provide the ability to acquire data quickly, provide access to specific areas of interest out of large masses of information, accommodate increasingly complex and evolving metadata, and be trusted to support long-term archiving.”
HDF5 datasets are either stored contiguously or in chunks which are indexed by a B-tree. The benefit of storing in chunks is that it allows for chunk-by-chunk compression. The downside is to access the data within a chunk, the entire chunk must be accessed.
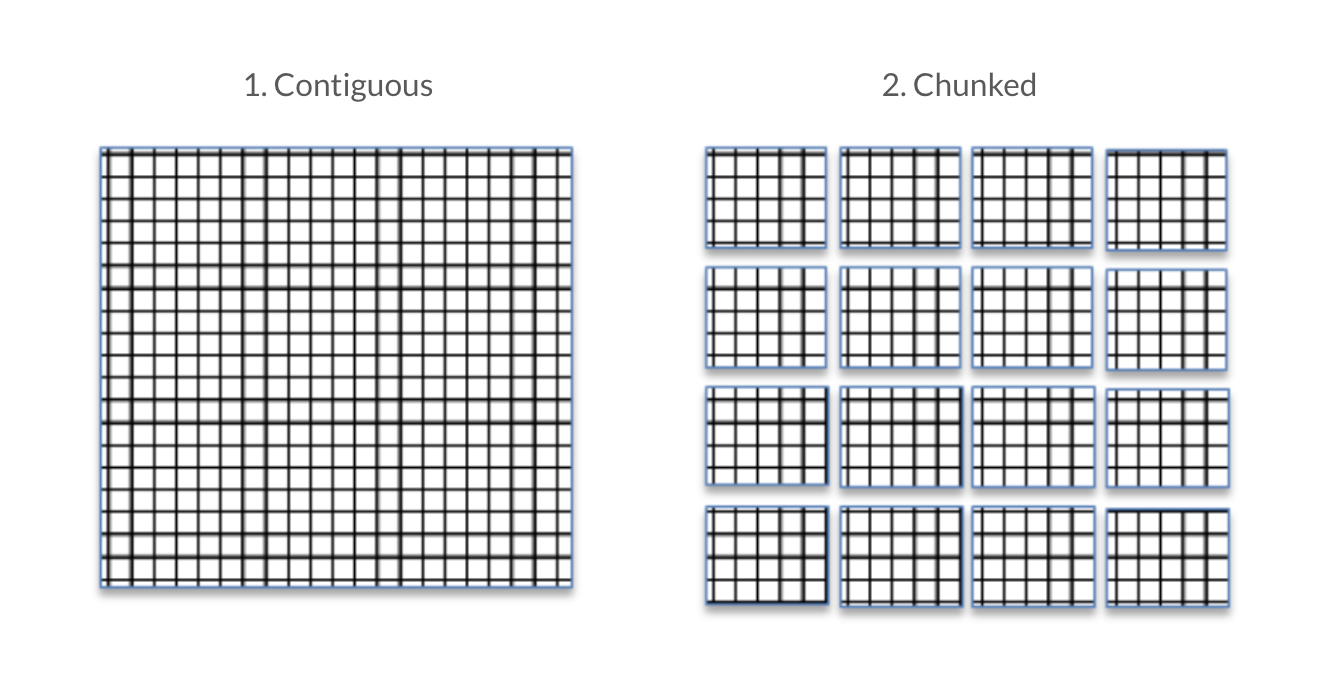
Source: Improving I/O Performance When Working with HDF5 Compressed Datasets
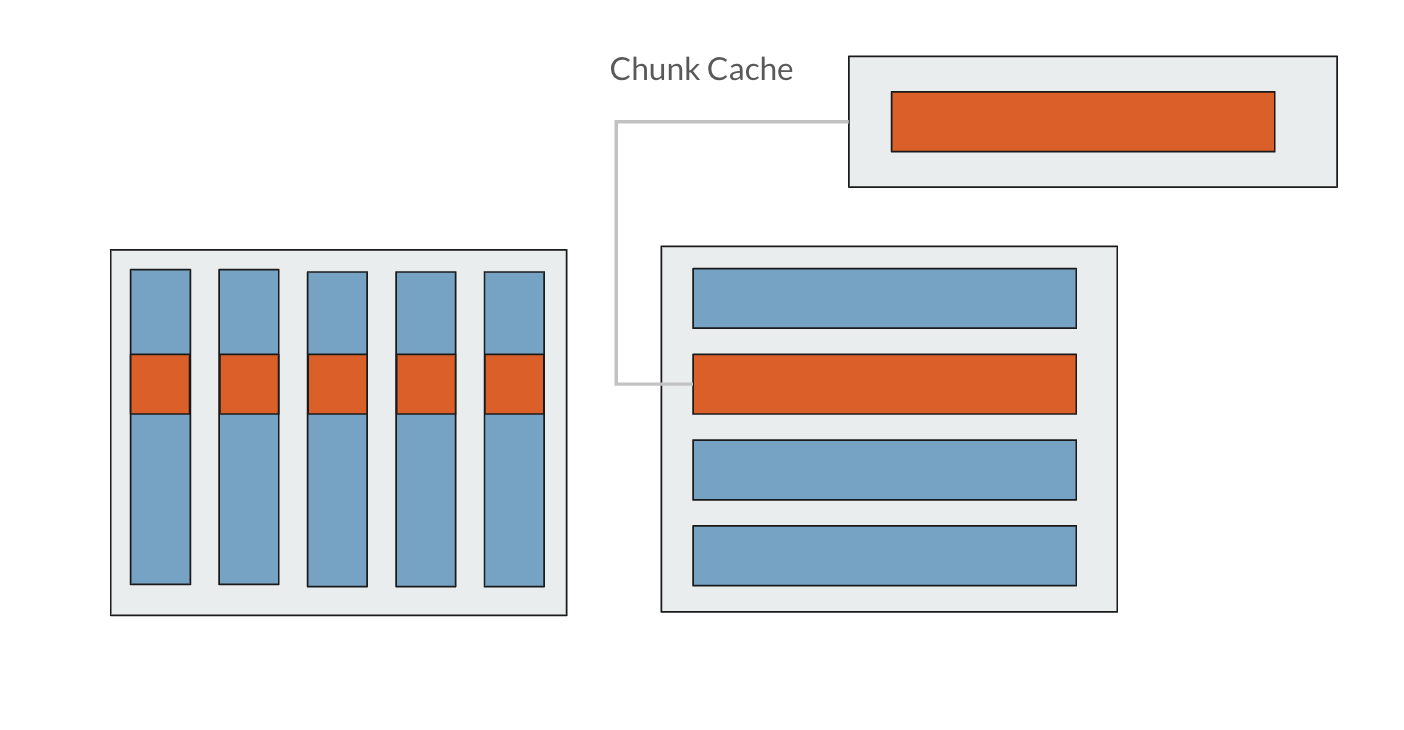
So to read just one element of a chunked dataset, the entire chunk must be read into the chunk cache, the entire chunk must be decompressed, and only then can the individual element be read by the application. Fortunately, once a chunk is in the chunk cache, any data within that chunk can be read by the application without needing to access disk and decompress again. However, as the chunk cache is only so large and can only fit so many chunks at a time, chunks will be kicked out of the cache once enough other chunks have been accessed. The default chunk cache size is 1MB, but it can be modified if your system has the resources. It is always a good idea to make a file’s chunk size smaller than the system’s chunk cache.
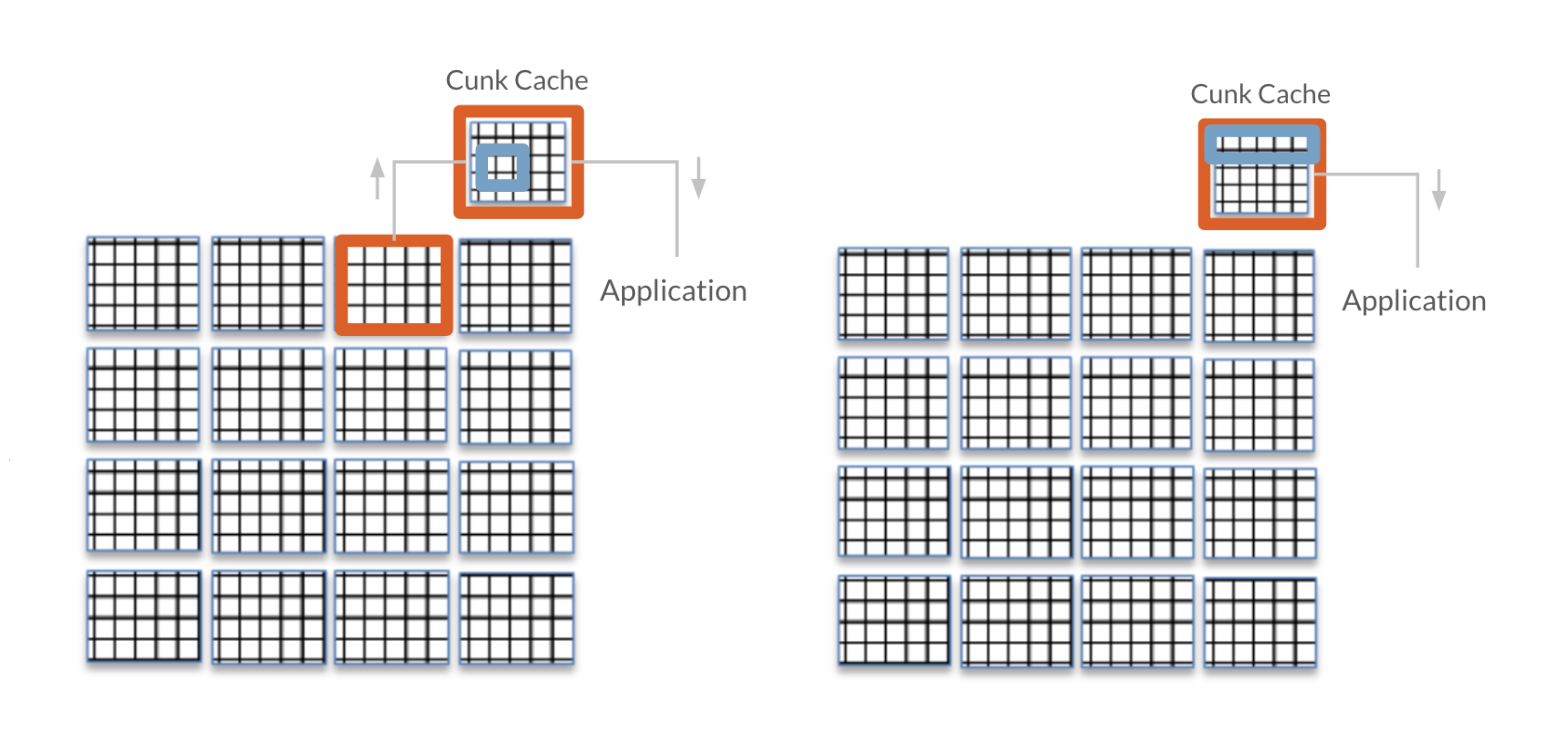
Source: Improving I/O Performance When Working with HDF5 Compressed Datasets with minor changes to illustrate the chunk cache
Selecting an appropriate chunk size is not as important as selecting an appropriate chunk dimension; if you are aware of the way in which your application will access the data within a dataset, you can align the chunks to the anticipated access pattern to improve access time. For example, given you have an AxB or (A, B) dimensional dataset, if you chunk by (1, B), then for each row from 0 to A, you have one chunk that contains all the columns, or 0 to B. If you chunk by (A, 1), for each column from 0 to B, you have one chunk that contains all the rows from 0 to A. This is the example illustrated below.
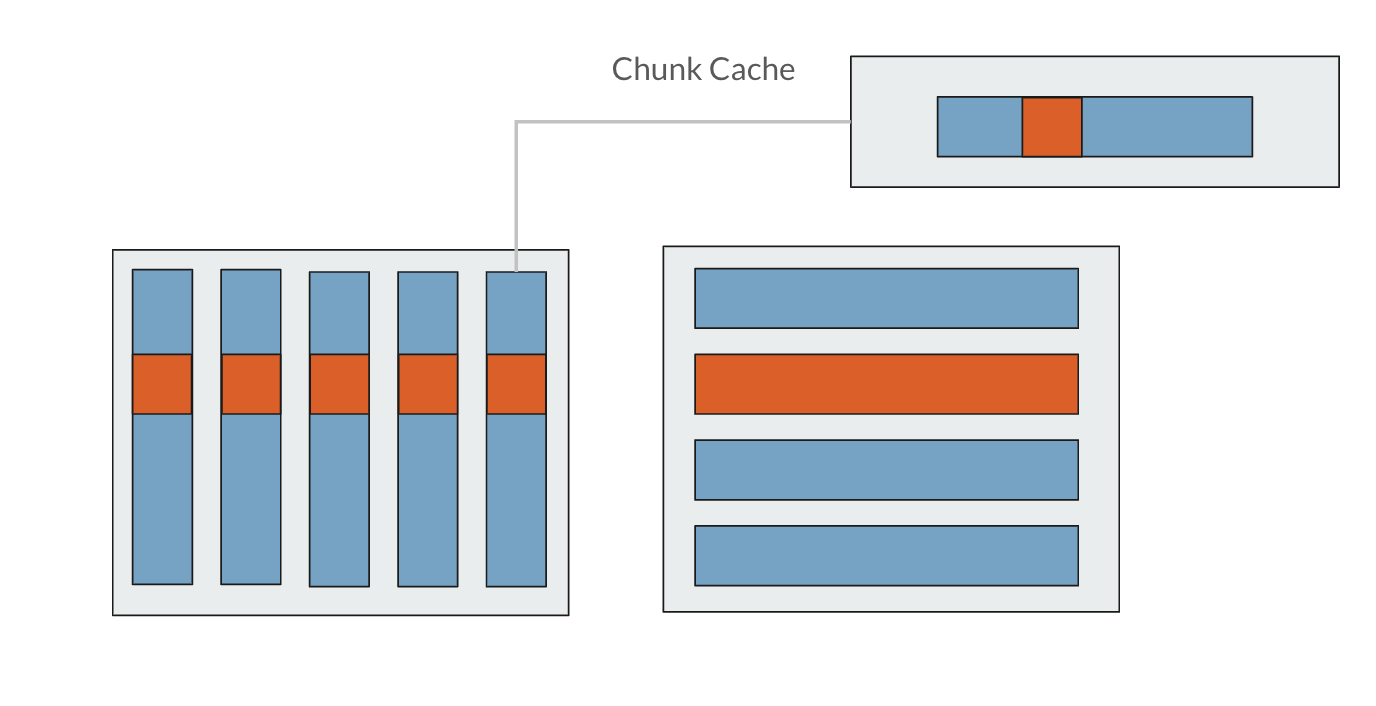
Say for this dataset with (A, 1) dimensional chunks, you want to access one row of data, seen in orange above. To access that small square of orange data in the chunk, you have to read the entire chunk into the chunk cache before you can read in the small portion of data that you actually want to access. So, to access just one row, you would actually have to read in the entire file from disk!
Contrast this with (1, B) dimensional chunks, seen above. Because the chunk dimension aligns with the rows, to access one row, you only have to read in one chunk of data. In this way, if you are knowledgeable about the access patterns that your file will need to accommodate, you can select a chunk dimensionality that aligns with it to improve access time. In a similar vein, if you are repeatedly accessing segments of data that are much smaller than your chunk sizes, chunking will have more of a detriment to your access time.
Tests
I created a couple Python scripts to test different chunking methods and the compression ratio and read and write times associated with each method on video files. To interface with the HDF5 files, I used h5py.
Analyzing HDF5 performance on video data
The chunk.py program converts a MOV or AVI video file into the other format with the Python package imageio, and runs tests on the read time of a single frame from that video.
Access time & file size with different chunking methods
The chunk.py program also converts a MOV or AVI video into HDF5 files with the following chunking methods:
- Default
h5pychunking method - One chunk containing the entire video
- One chunk per frame in the video
- One chunk per frame and color channel in the video
The dimensions of the datasets read from the video files are as follows: (number of frames, height of frame, width of frame, color channel).
It also converts the video to an uncompressed, contiguous (non-chunked) HDF5 file for comparison. Running the chunk.py program with the -t flag runs the access time tests, which analyze the following access patterns:
- Reading and writing one pixel within the video
- Reading and writing one frame within the video
- Reading and writing the entire video
To ensure the reading and writing operations were timed as independent operations, the file under test was closed and reopened between each read or write, and the chunk cache size was set to 0 bytes. The file was also reopened for each test; calling close() on a h5py is equivalent to a setting the close degree to H5F_CLOSE_STRONG, i.e. closing all open objects within the file, such as datasets or groups that were being written or read from.
# test read time of one pixel
# rdcc_nbytes = chunk cache size
f = h5py.File(path, 'r+', rdcc_nbytes=0)
start = time.time()
# frame number, height, width, color channel
val = f[dset_name][27,0,0,0]
end = time.time()
f.close()
print(f'Pixel read time: {end-start}')
# test write time of one pixel
# rdcc_nbytes = chunk cache size
f = h5py.File(path, 'r+', rdcc_nbytes=0)
start = time.time()
# frame number, height, width, color channel
f[dset_name][27,0,0,0] = 0
end = time.time()
f.close()
print(f'Pixel write time: {end-start}')
Results
HDF5, MOV, and AVI
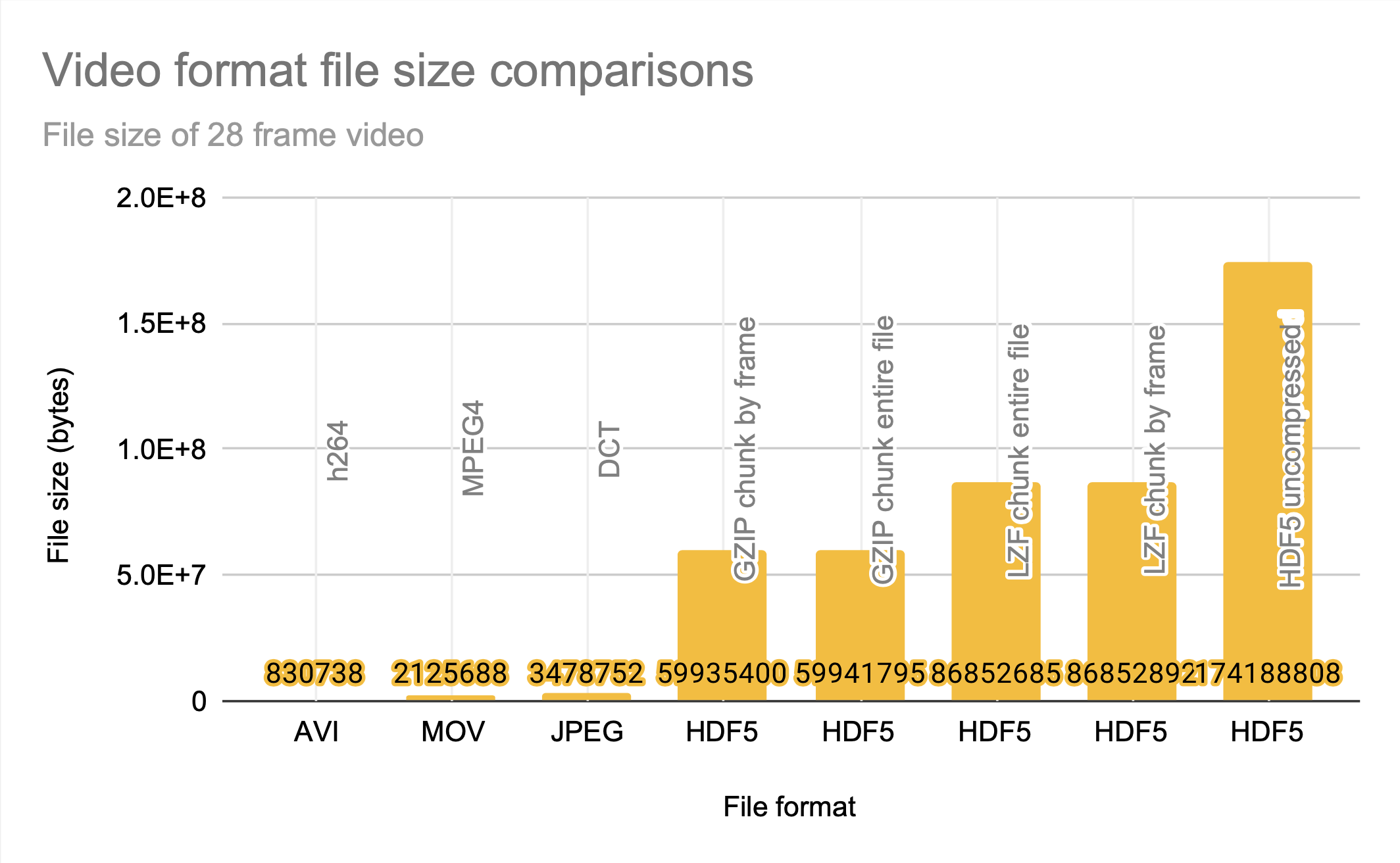
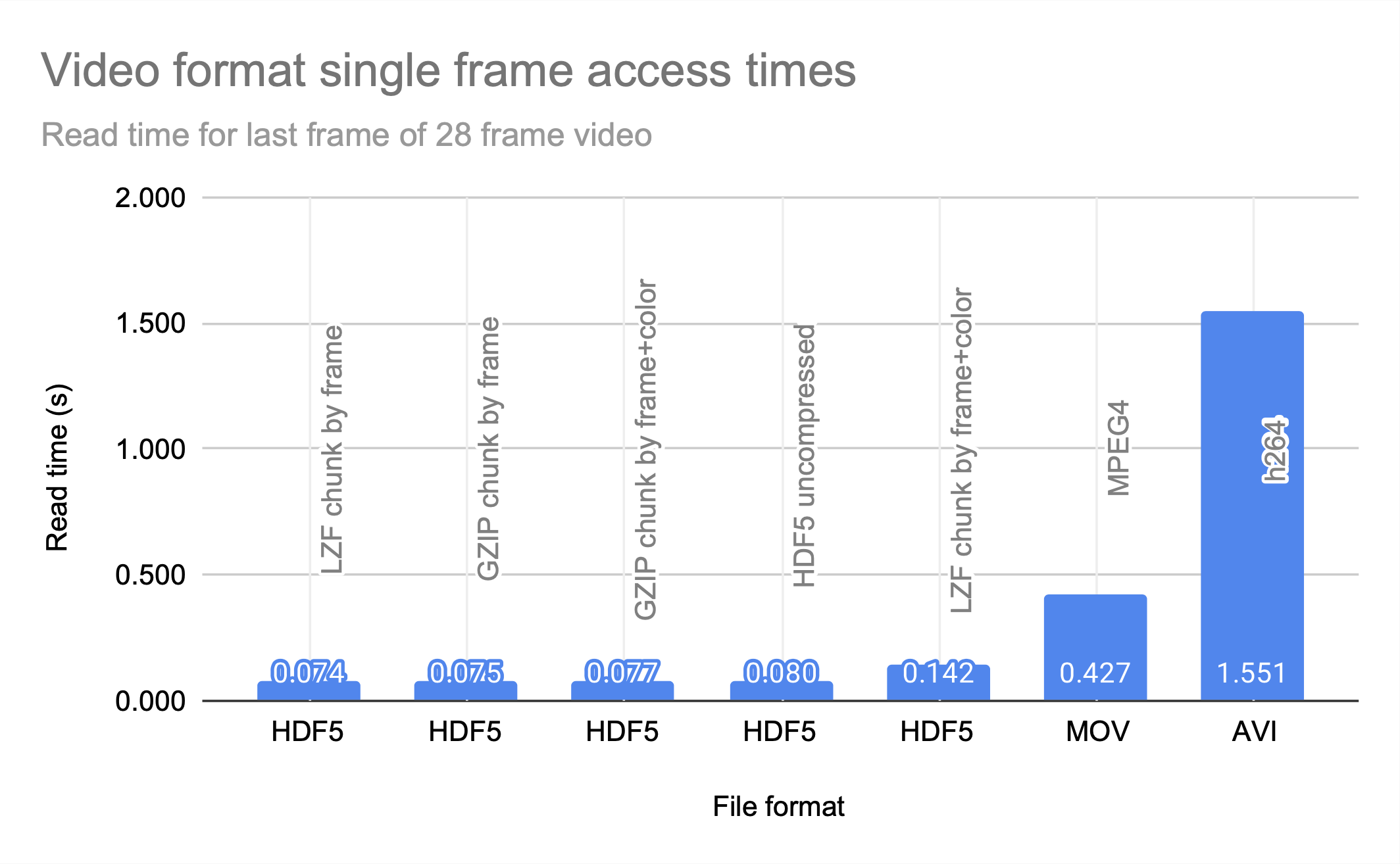
HDF5 clearly prioritizes access time over compression, and is a good choice for applications that are knowledgeable about their access patterns and can align the chunks to match them. The JPEG file size entry is the size of the sum of all 28 frames of the video stored as JPEG photos; even without motion-prediction coding, the HDF5 files are significantly less space efficient.
Chunking methods
Compression ratio achieved with gzip for different chunking methods has a general trend of a smaller chunk size leading to larger files; more chunks mean more entries into the B-tree the chunks are stored in, and smaller intervals that can be compressed.
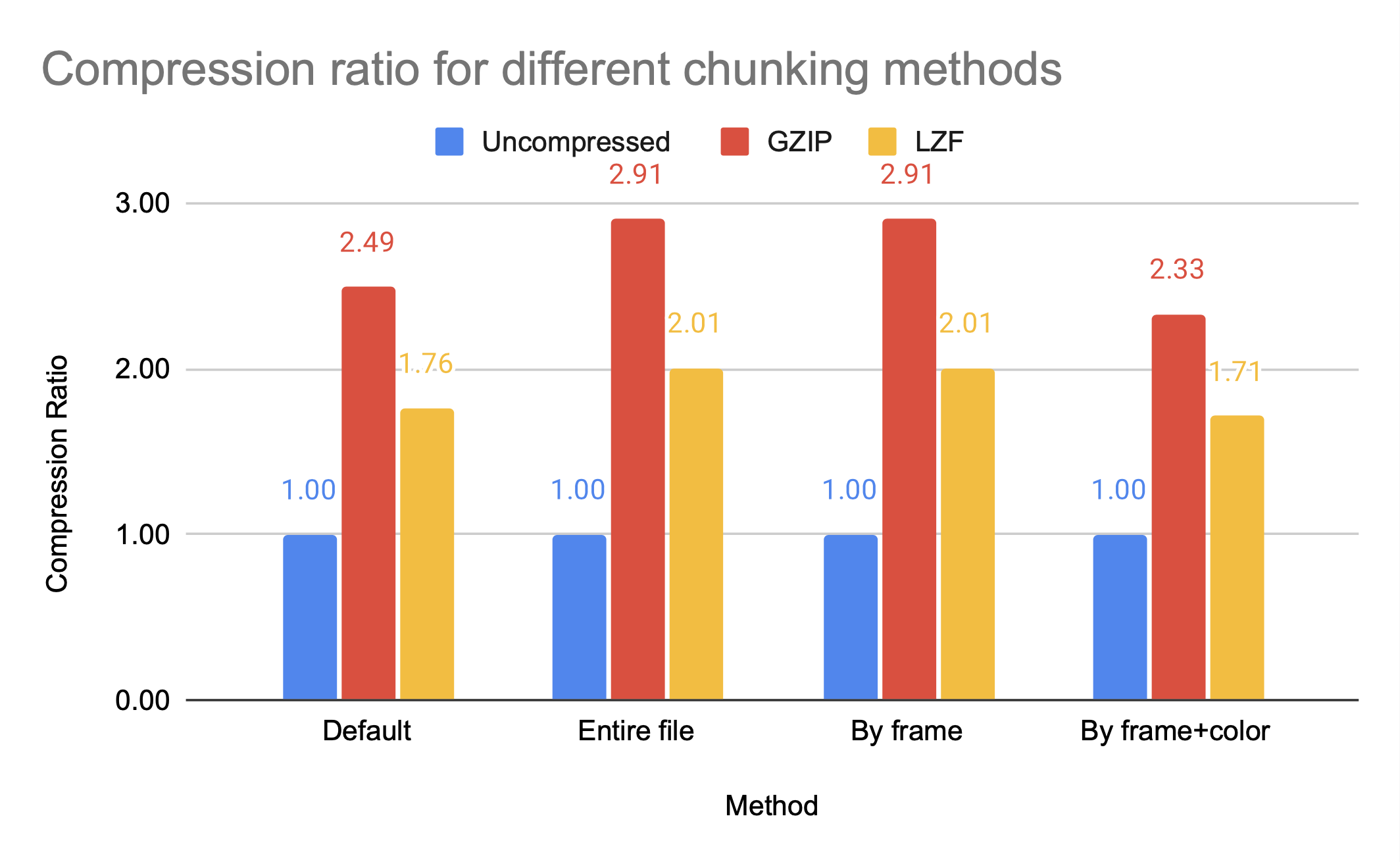
The graph above shows that the chunking methods whole and by frame, which are larger chunks, have higher compression ratios than the smaller chunk by frame+color. However, by frame+color has significantly faster access times, as seen in the figures below, which show the read and write times for HDF5 video files with different chunking methods, all compressed with gzip.
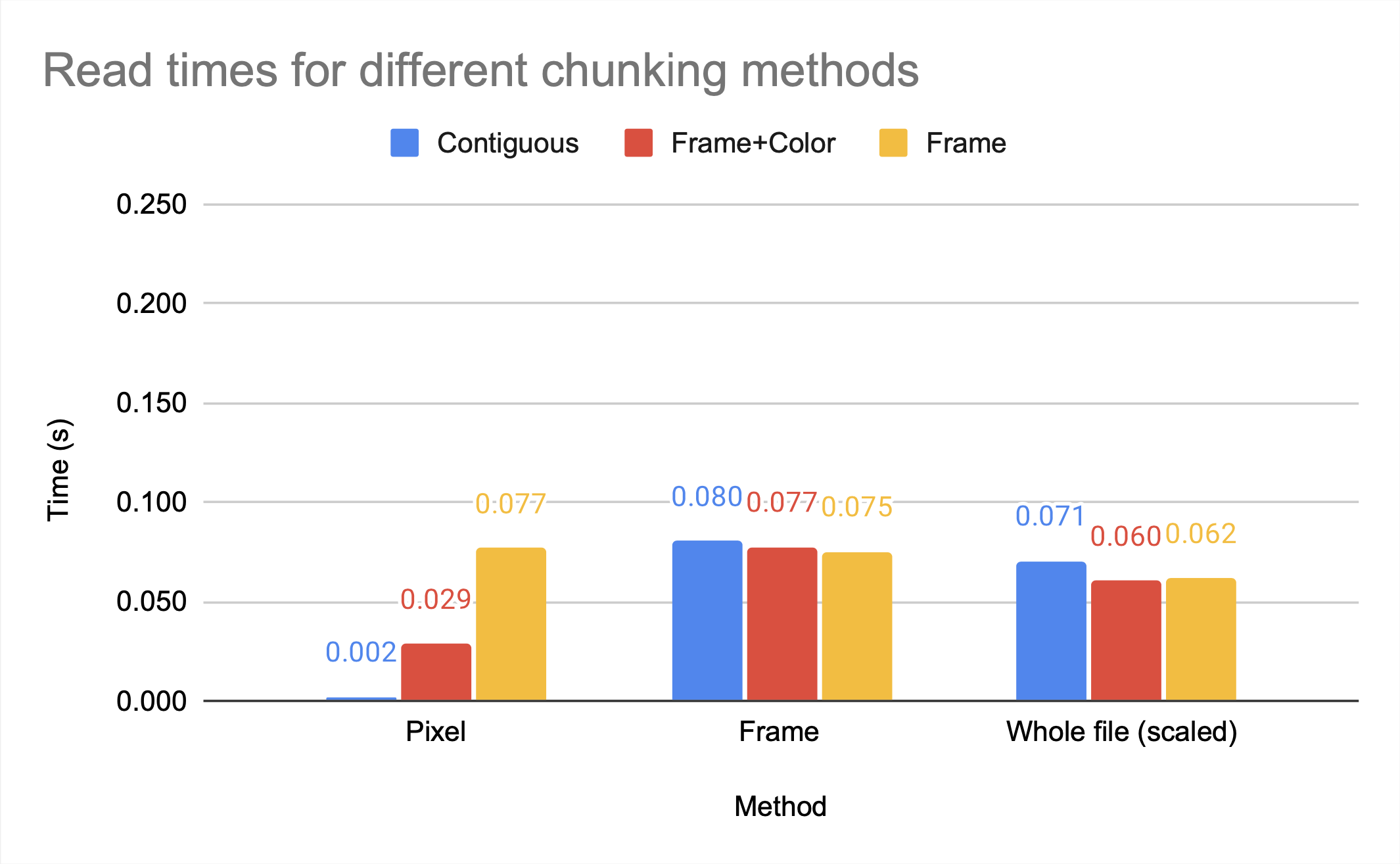
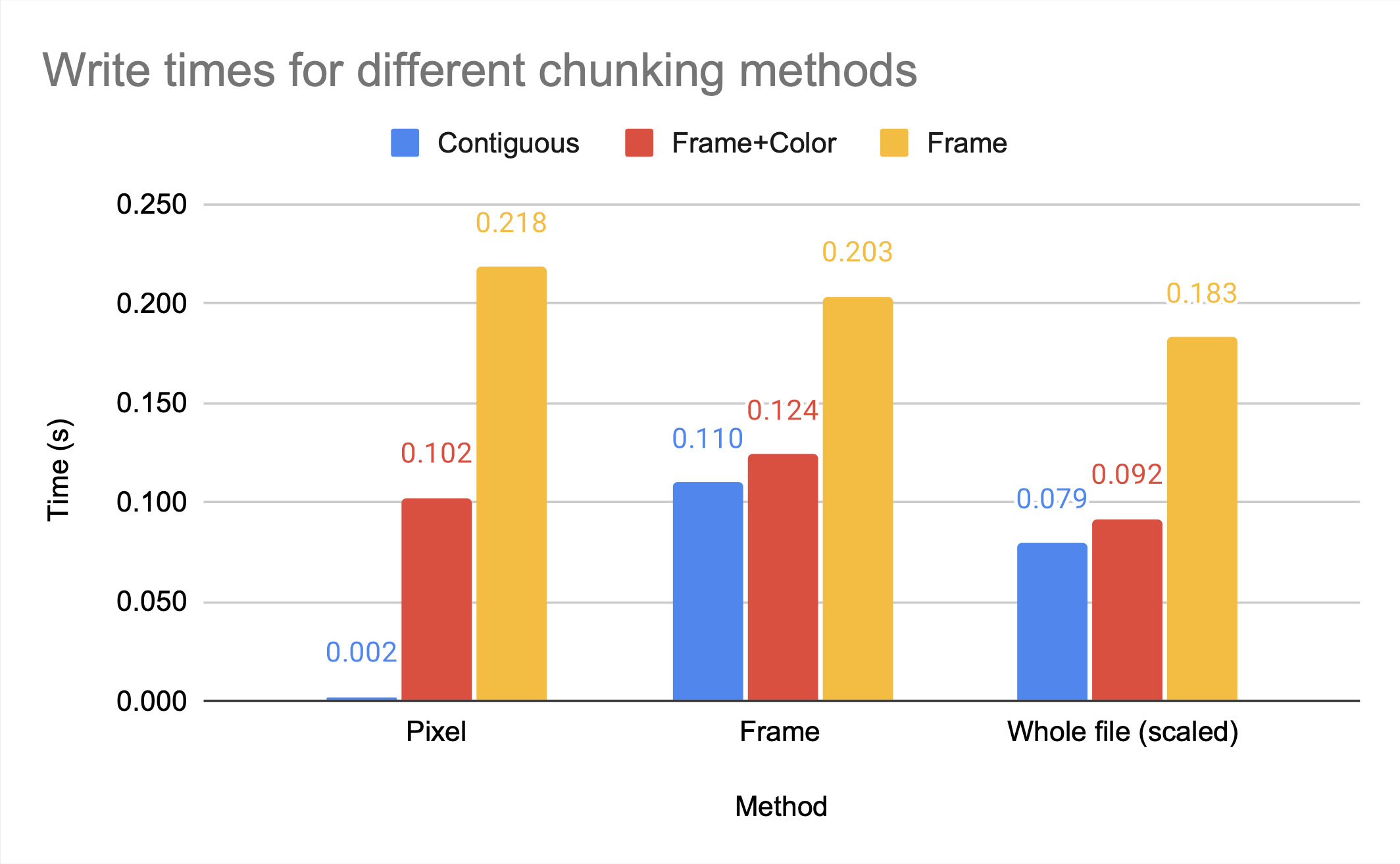
Note that gzip compression scales poorly, which is the cause of the inefficient write times for by frame but not by frame+color. While whole and by frame provide similar compression ratios, the access time to the whole file is significantly worse than the access time to the by frame file unless the entire file is being accessed.
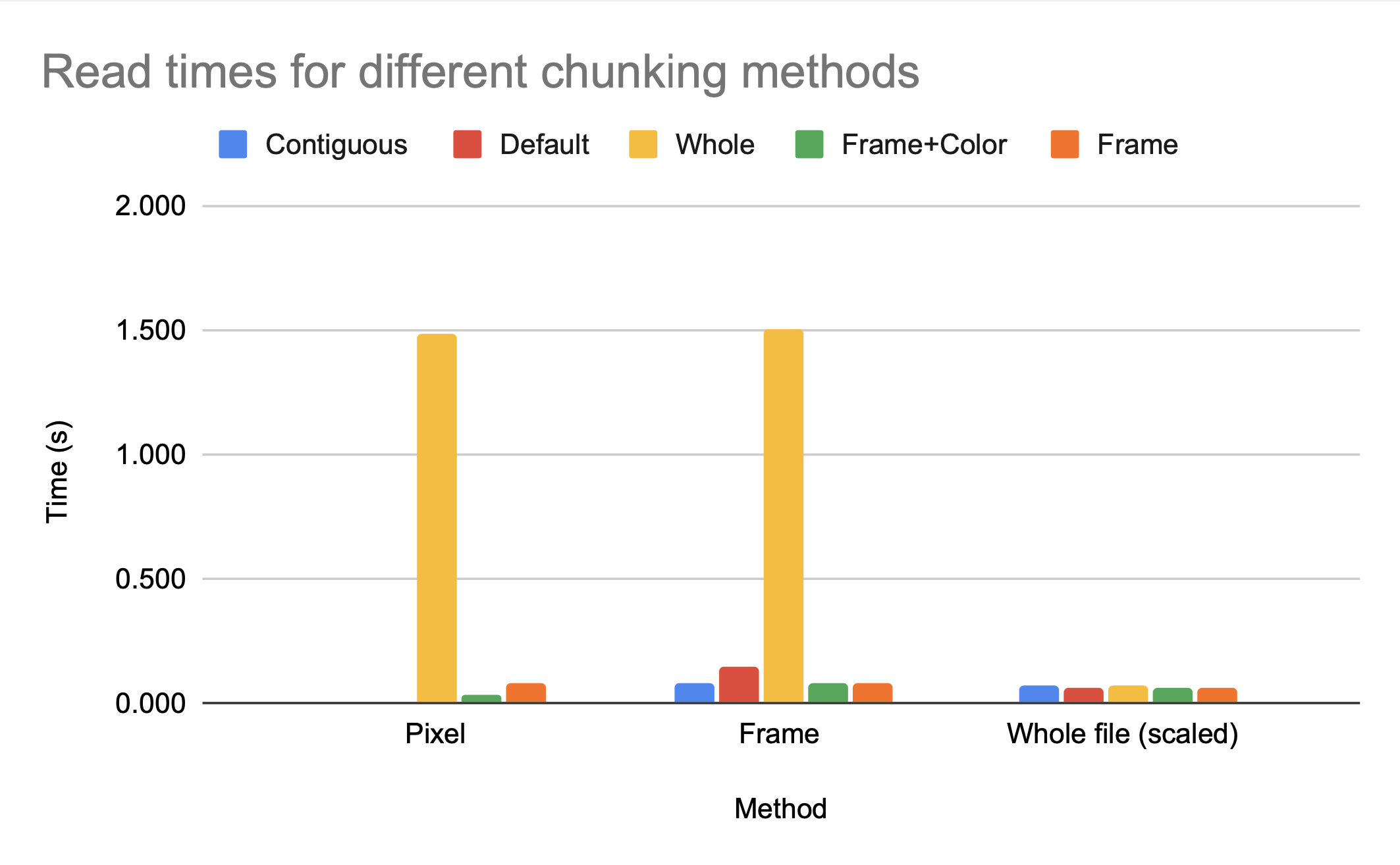
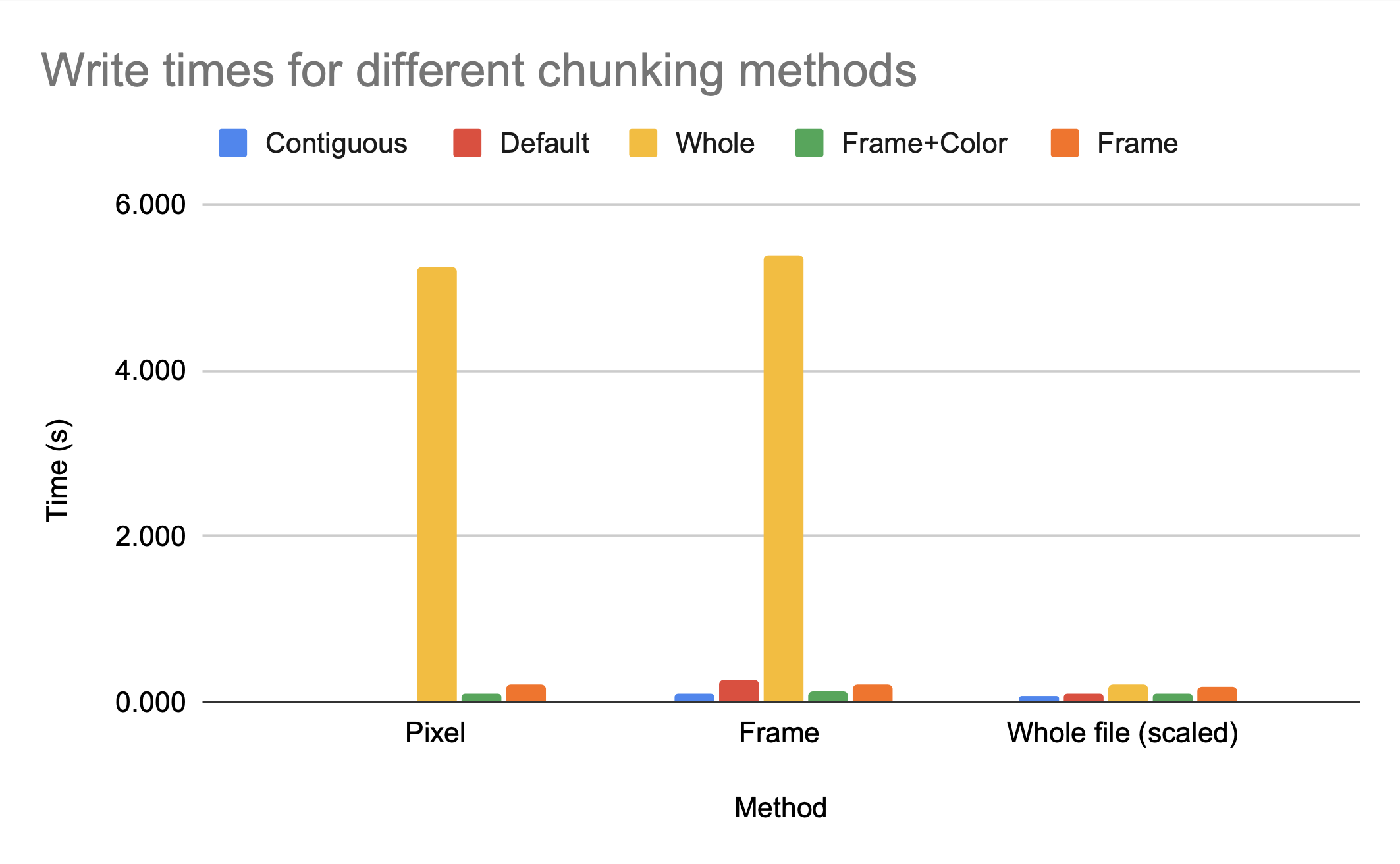
This makes logical sense; if the access is an access to the entire file, then every chunk will have to be accessed, compressed, and/or uncompressed regardless of the chunk size. If you are making an application where the access pattern is known to be the entire file, you can optimize chunk size for compression with relatively little cost to access time.
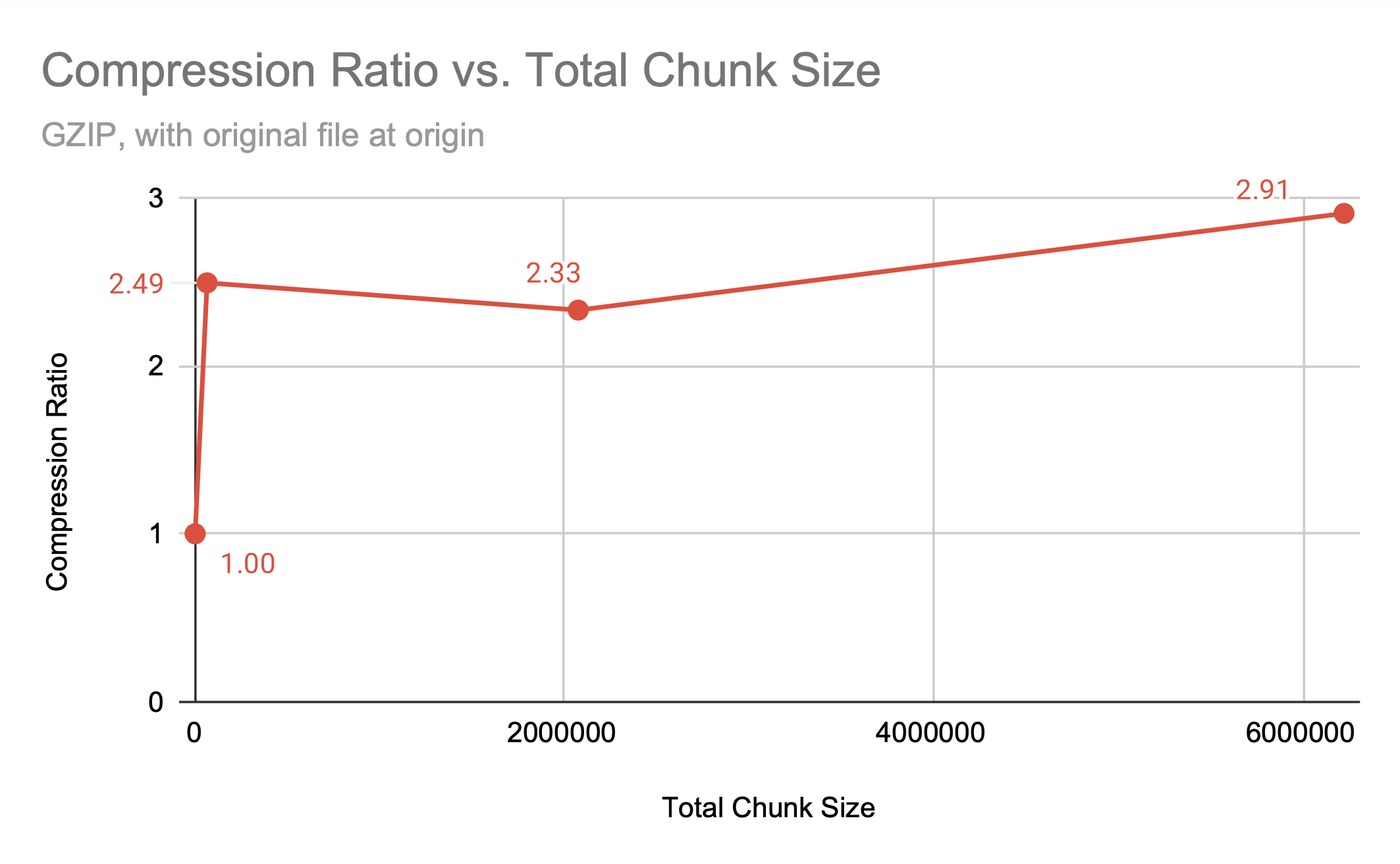
Note that the better compression ratio is not a result of the larger chunk size; the default h5py chunk dimensions provide a better compression ratio than chunking by frame+color, despite having significantly more chunks of a smaller size. While there is a relationship between chunk size and compression ratio, it is clearly not the only factor that determines how space efficient the file can be.
| Method | Chunk size (bytes) | Number of chunks | Dimensions | Compression Ratio |
|---|---|---|---|---|
| h5py default | 64800 | 2688 | (2, 240, 135, 1) | 2.4931 |
| by frame+color | 2073600 | 84 | (1, 1920, 1080, 1) | 2.3309 |
However, because we know the access pattern is by frame, we know the default chunking method will not provide efficient access time; the dimensionality is (2, 240, 135, 1), so multiple frames are grouped together but the pixels for each frame are split among blocks. Accessing one frame would involve decompressing multiple chunks.
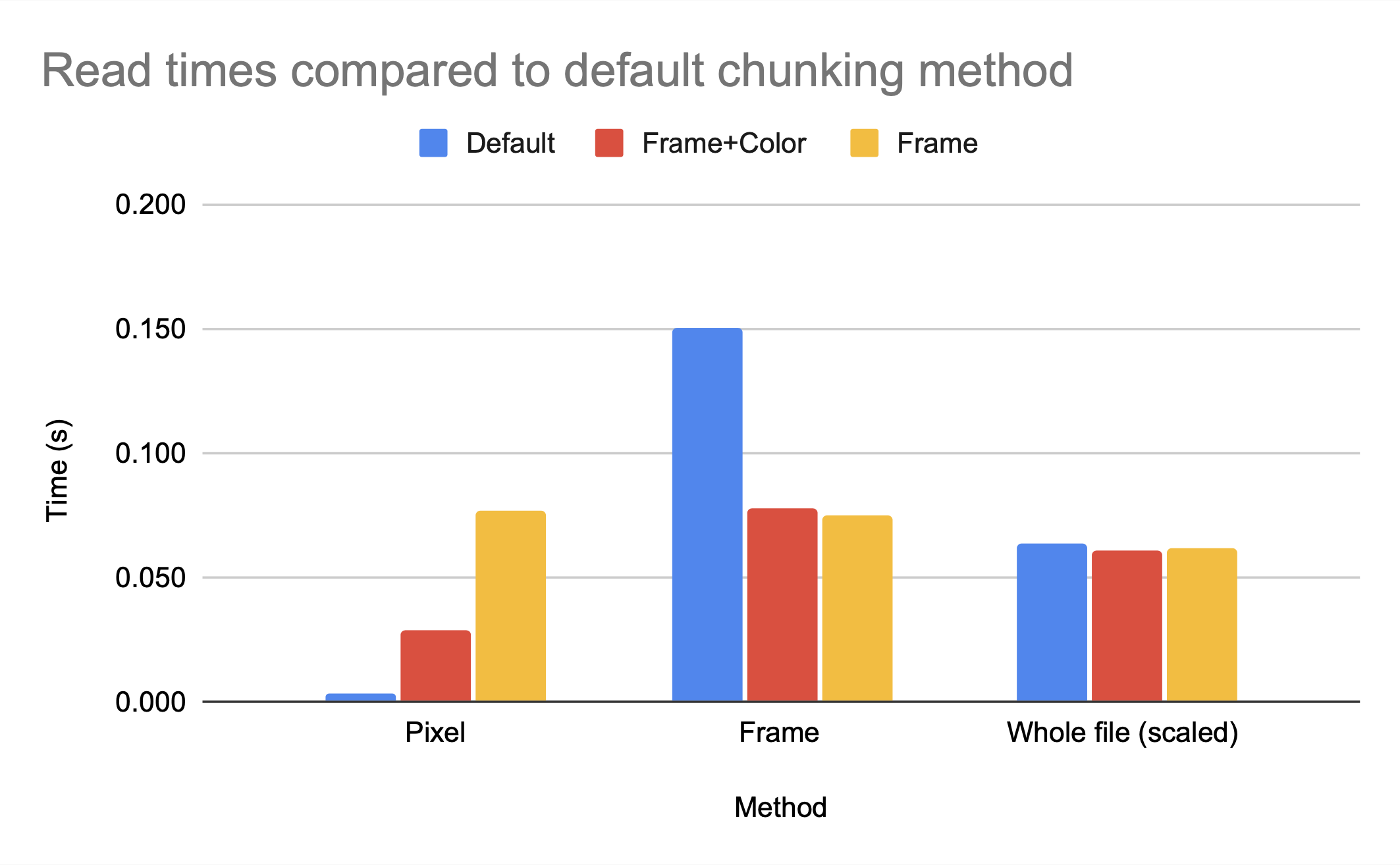
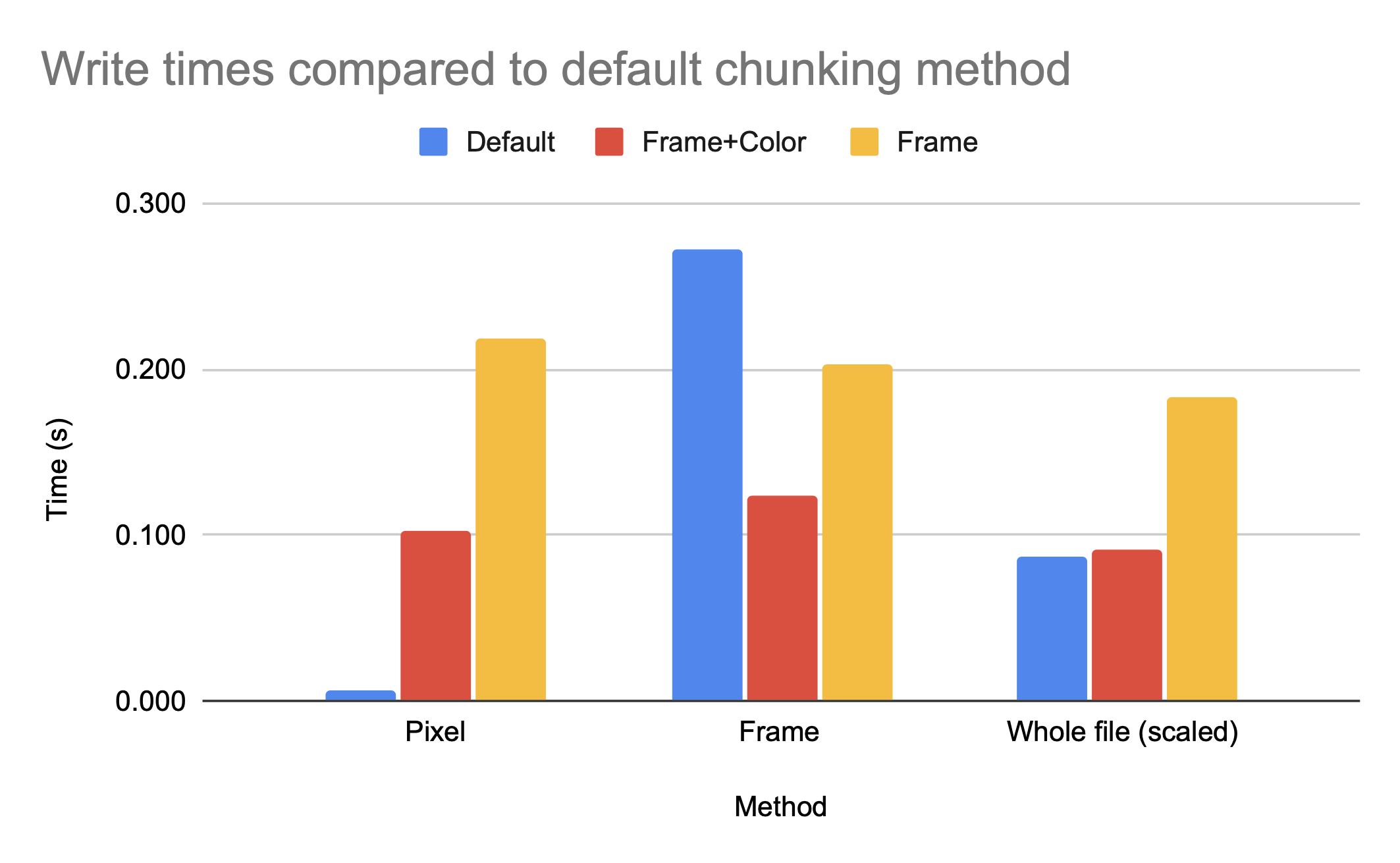
While the access time for a single pixel is clearly much better for default, which makes sense as default has smaller chunk sizes and therefore less data has to be read in per element accessed, the access time for a single frame is significantly worse for default then for either by frame+color or by frame. While the h5py python package can make intelligent decisions on chunk sizes for compression ratio, only someone with knowledge of the anticipated access patterns can make an intelligent decision on the chunk dimensions.
Compression algorithms
The available 3rd-party compression algorithms from hdf5plugin included algorithms that performed better than and worse than the HDF5-included gzip when it came to total compression achieved.
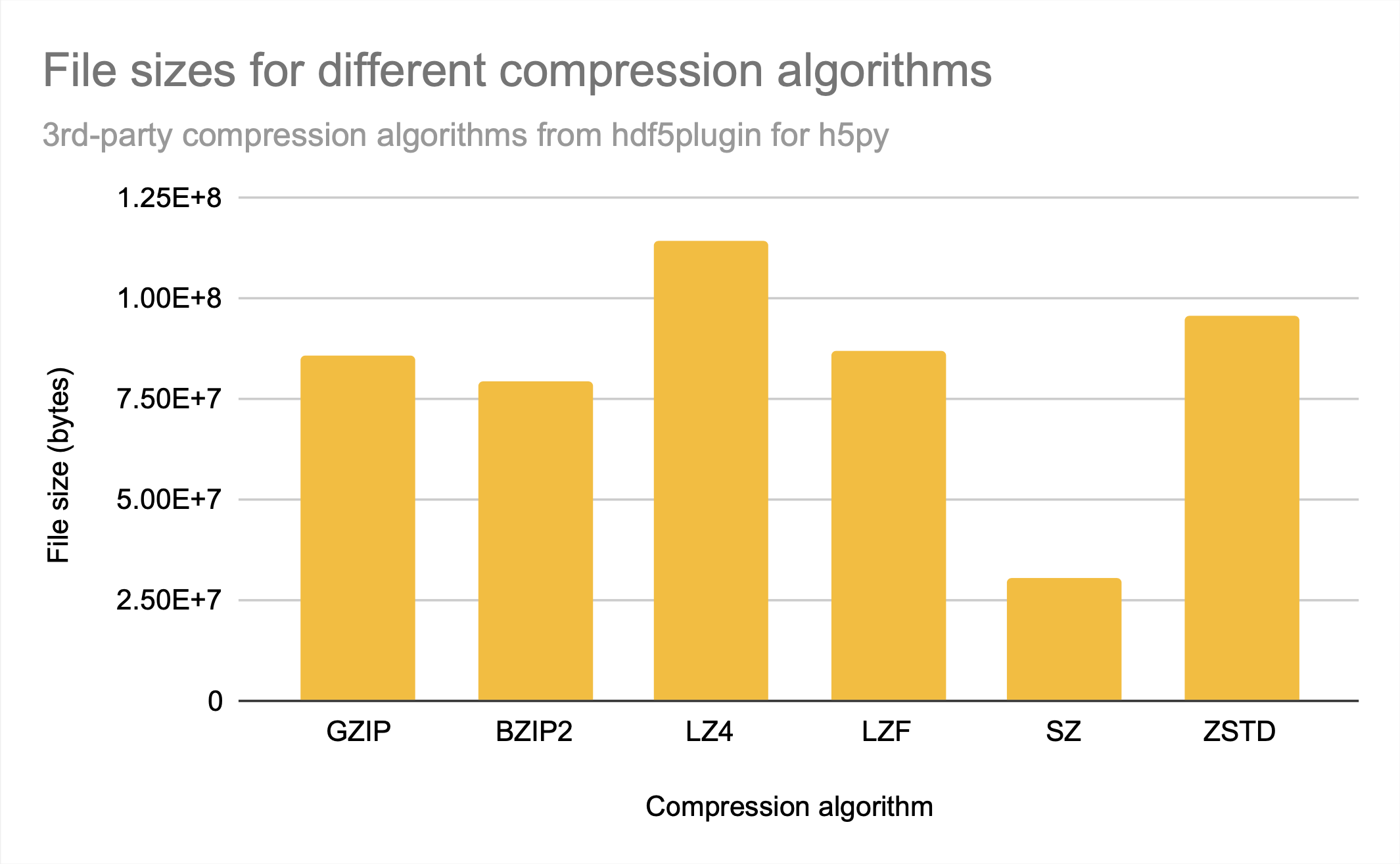
While sz is clearly the best compression algorithm among those provided for file size, lz4 and zstd had the fastest access times. While they provide similar access times, zstd has a bit better compression ratio.
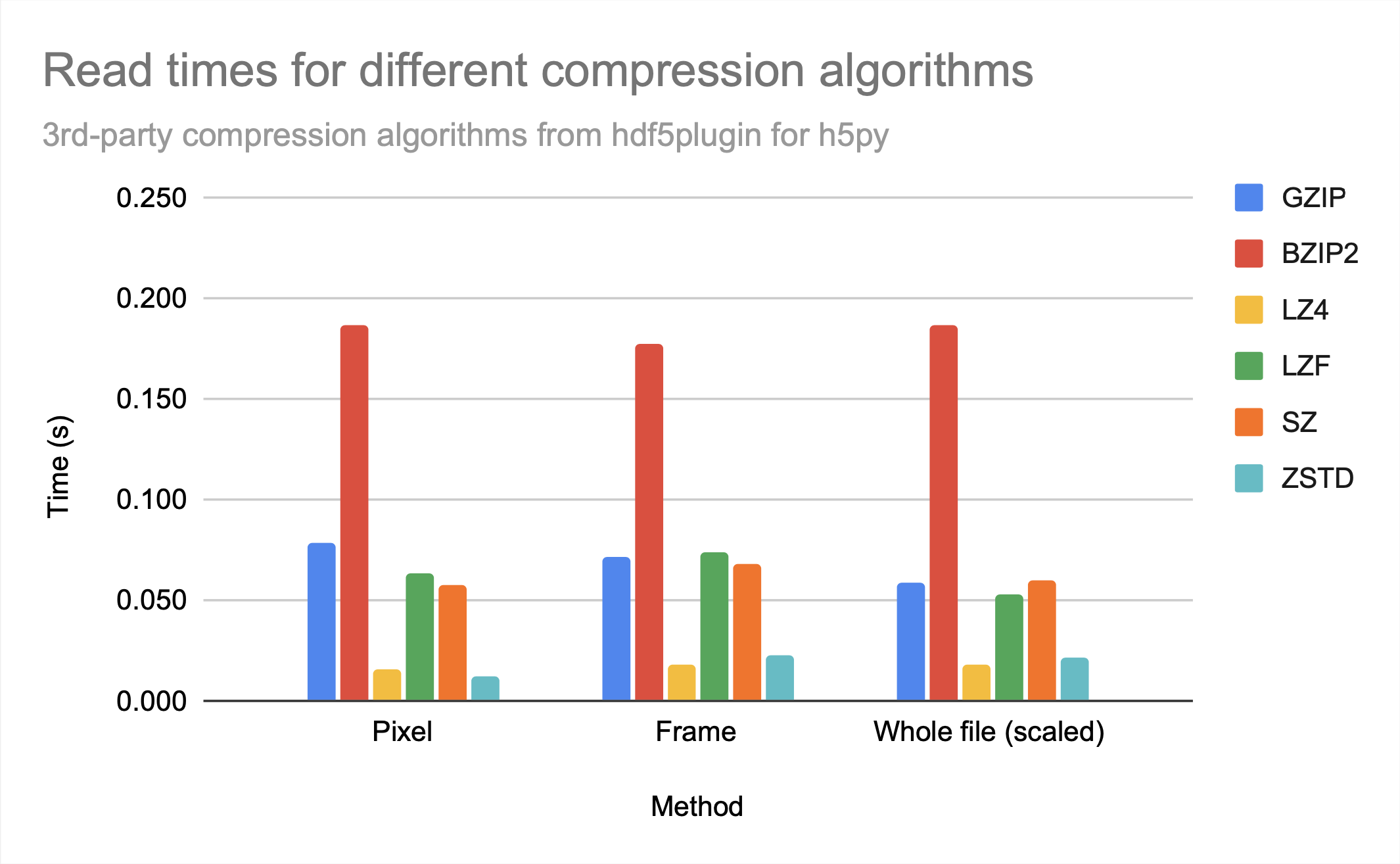
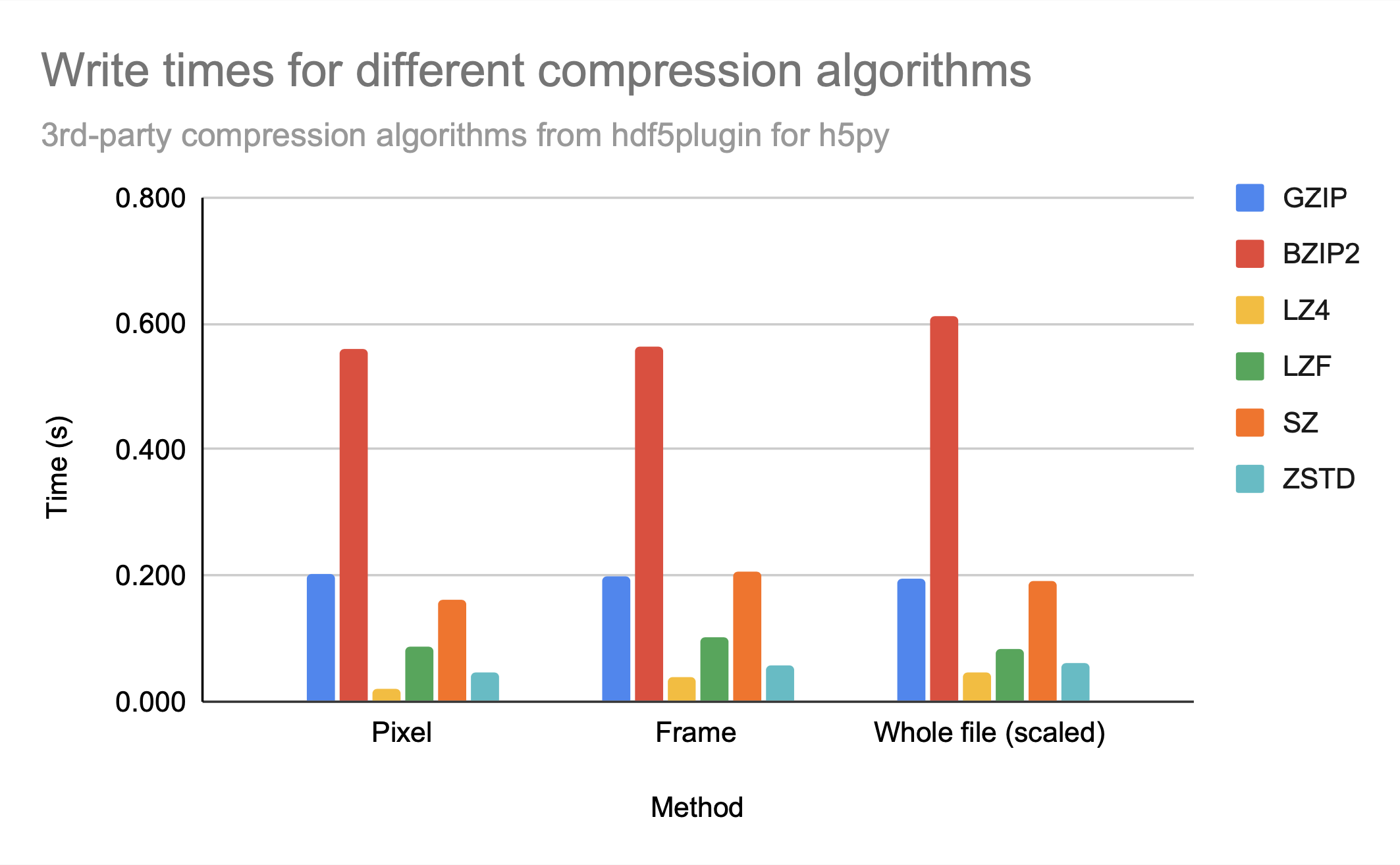
Note that bzip2 seems to provide neither relatively good compression nor relatively fast access times in comparison to other available 3rd-party compression algorithms, as gzip, which creates only a slightly larger file, has significantly better access times, similar to sz.
For all the compression algorithms provided, read time is worse than write time, probably indicating that compression takes longer than decompression.
Conclusions
HDF5files prioritizespeedof access overcompression- You can avoid detriment to access time by aligning your chunks with your access pattern; because
chunks are stored contiguously, accessing a chunk only costs the compression and decompression time - Aside from selecting a chunk size that strikes a balance between compression and access time that best fits your application, you must also
select a compression algorithmthat strikes a balance between compression and decompression that best fits your application - If you really only want to access the entire file at a time, then compression is
(almost)free
Run these tests yourself
- Choose a
MOVorAVIvideo (<= 40 frames recommended as it is processing intensive) - Clone the
github - Install
Python 3.10& check version to ensure 3.10 is installed:python3 --version - Run
./install.shto locally installpip3packages required for this project (or viewinstall.shand install the packages manually) - Make a
files/directory and copy your selected video into it - Run
./chunks.py -f files/[your_file.mov]to convert your video toHDF5files - Modify the dimensions of the I/O test as shown below
- Run
./chunks.py -f files/[your_file.mov] -tto run the I/O tests
Note: the dimensions within the access time test may need to be modified, as the current dimensions are hardcoded to the dimensions of my test file; (28, 1920, 1080, 3), or a 28-frame 1920x1080 pixel 3-channel color video.
# test read of entire file
f = h5py.File(path, 'r+', rdcc_nbytes=0)
start = time.time()
frame = f[dset_name][0:27,0:1919,0:1079,0:2]
end = time.time()
f[dset_name][0:27,0:1919,0:1079,0:2] = frame
f.close()
The slices to read the entire file are from 0:number_of_frames-1, 0:height_of_frame-1, 0:width_of_frame-1, and 0:number_of_color_channels-1. For example, to modify this test to run for a 30-frame 3072x2304 color video, i.e. a video with dimensions (30, 3072, 2304, 3), the modified code would be:
# test read of entire file
f = h5py.File(path, 'r+', rdcc_nbytes=0)
start = time.time()
frame = f[dset_name][0:29,0:3071,0:2303,0:2]
end = time.time()
f[dset_name][0:29,0:3071,0:2303,0:2] = frame
f.close()
The dimensions of your selected video’s frames are displayed when running ./chunks.py, after displaying the metadata on that video. For example, my video has a frame dimension of (1920, 1080, 3).
Path: files/movies/katrina.mov
Size: 2125688
...
'rotate': 0,
'size': (1080, 1920),
'source_size': (1080, 1920)}
Shape: (1920, 1080, 3)
The number of frames in your selected video can be discovered by running ./chunks.py -j to write the frames as JPEG images. For example, my video has 28 frames.
Writing JPEGs...
Frame 1: 127850
Frame 2: 129607
...
Frame 28: 126611
Saved frames 1-28
Total size: 3478752
Since the dimensions are (number of frames, height of frame, width of frame, number of color channels), my total video dimensions are: (28, 1920, 1080, 3), so my slices are as follows in the I/O test:
# last frame's 0x0 pixel on color channel 0
pixel = f[dset_name][27,0,0,0]
# last frame's 1920x1080 image on all three channels
one_frame = f[dset_name][27,0:1919,0:1079,0:2]
# all frames of all 1920x1080 images on all three channels
all_frames = f[dset_name][0:29,0:3071,0:2303,0:2]
Or run some new experiments
Third-party compression algorithms
Multiple compression algorithms are available for the chunked HDF5 files, including 3rd party compression algorithms. These can be specified with the -c [algorithm] option. While I compared the differences in file sizes and access time for different types of access for most of these algorithms when chunking by frame, a future experiment could be comparing compression algorithms’ performance with different chunking methods or different data types.
# Available compression algorithms & their names
ALGS = {'gzip' : 'gzip',
'lzf' : 'lzf',
'bitshuffle': hdf5plugin.Bitshuffle(),
'blosc' : hdf5plugin.Blosc(),
'bzip2' : hdf5plugin.BZip2(),
'lz4' : hdf5plugin.LZ4(),
'sz' : hdf5plugin.SZ(absolute=0.1),
'zfp' : hdf5plugin.Zfp(reversible=True),
'zstd' : hdf5plugin.Zstd()}
Some additional, but unanalyzed results are in the results directory.
Chunk cache
I removed the chunk cache for my experiments, which in many cases significantly speeds up access time for chunked files. Future work could include what types of access patterns most benefit from the chunk cache or what factors should be considered when deciding the chunk cache size.
Memory access type
Memory accesses can be to the computer’s primary memory, like RAM or ROM, or secondary memory, like the computer’s hard disk. A future experiment could involve comparing hard disk (HDD) access to other types of secondary memory (such as SSD).
Even when running experiments on access time for one type of secondary memory, it’s complicated. Operating systems present an illusion of contiguous memory, while allocating these blocks in memory to align with easier-to-access blocks in physical memory. Most also wait to write changes recorded in RAM to the HDD until the opportune moment and make replicas of files to protect against data loss. These, along with many other factors, play a role in measured access times.
Mixed datasets
One of the main benefits of HDF5 files is that they can store arbitrarily complex data types within the datasets, including complex data types, such as structs in C or classes in C++. A future experiment could involve comparing access time and compression ratio of HDF5 files and other file formats for increasingly complex data types to see if HDF5 provides any significant advantage over alternatives, or seeing how HDF5 compares to other mixed-type dataset file formats.
References
“6 hours of debugging can save you 5 minutes of reading documentation” - @jcsrb on Twitter
“I checked this out weeks ago for a bit of light reading” - Hermione
- Working with Hierarchical Data Format (HDF5) Files
- Chapter 4. How Chunking and Compression Can Help You from Python and HDF5 by Andrew Collette
- Earth Sciences: Why HDF Technologies?